One enzyme uses either guanine or hypoxanthine (adenine with the amino group replaced by an OH). A second enzyme uses free adenine. A third enzyme is specific for uracil and thymine. All the enzymes carry out the same reaction: transfer of the free base to the ribose‐5′‐monophosphate of PRPP, forming a nucleoside‐5′‐monophosphate (NMP).
Purine biosynthesis
Purine synthesis uses a PRPP “handle” where the ring is assembled to make a 5′ NMP, inosine monophosphate (IMP).

![]()
IMP is the common intermediate in purine biosynthesis, and can be converted to GMP or AMP as needed.
The first reaction in purine biosynthesis is the transfer of the amide from glutamine to PRPP with release of pyrophosphate. The product is phosphoribosylamine (PRA).

![]()
Then the amino acid glycine is transferred to PRA, making glycinamide mononucleotide.
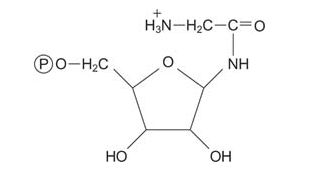
![]()
The amino group of glycine is formylated, with the formyl group being donated by N 10‐formyl‐tetrahydrofolate.
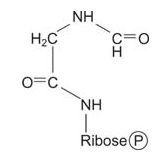
![]()
Now the amino NH 2 is transferred to the carboxyl carbon of glycine from glutamin, with ATP as an energy source. This compound, formylglycineamidine ribonucleotide, closes to make the “smaller” (imidazole) ring of the purine. Again, ring closure uses ATP energy.
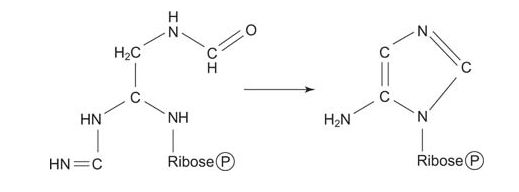
![]()
Now the larger ring is built on the smaller one. A carboxylation reaction with CO 2 starts synthesis of the 6‐membered ring.
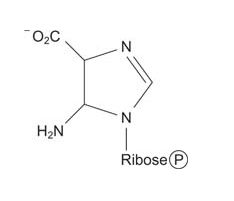
![]()
Then the amino group of aspartate is transferred to the carboxyl, making an amide. This condensation uses ATP and the amide is cleaved to release fumarate, leaving behind the imidazole with a 5‐amino group (left from the amidation of glycine four steps earlier) and a 4‐carboxamide. (Note how this reaction is similar to the formation of arginine during the urea cycle.)
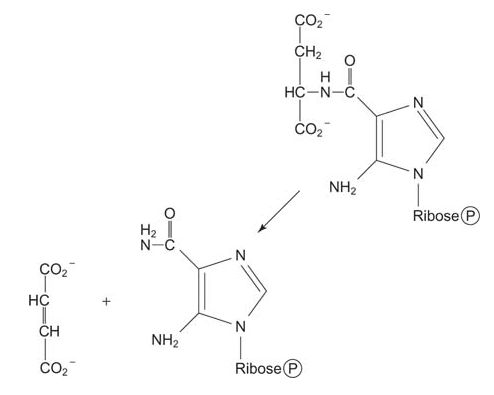
![]()
Eight of the nine components of the ring are now present. The last ring component comes from a 1‐carbon transfer of a formyl group from N 10‐formyltetrahydrofolate.

![]()
Finally, the ring is closed by dehydration to yield IMP.
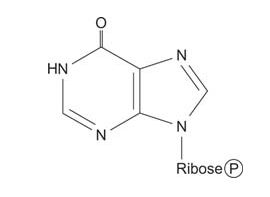
![]()
IMP is the key intermediate of purine nucleotide biosynthesis. IMP can react along two pathways that yield either GMP or AMP. Oxidation of the 2 position makes xanthine monophosphate, which is transamidated to GMP. Alternatively, the α‐amino group of aspartate can replace the ring oxygen of IMP to make AMP. (Note again how this reaction is similar to the synthesis of arginine from citrulline.)

The rates of these two complementary reactions can control the amount of either AMP or GMP present in the cell. Each of these reactions is feedback‐inhibited by its nucleotide product. Thus, if more adenosine nucleotides exist than guanosine nucleotides, the synthesis of AMP slows down until the purine nucleotides balance.
Degradation of purine nucleotides
Extra purines in the diet must be eliminated. In mammals, the product of purine breakdown is a weak acid, uric acid, which is a purine with oxygen at each of three carbons.
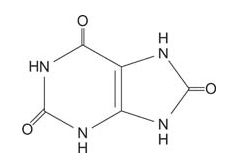
![]()
Uric acid is the major nitrogen excretion product in birds and reptiles, where it is responsible for the white, chalky appearance of these droppings. Uric acid is poorly soluble in water, and in humans, formation of uric acid crystals is responsible for the painful symptoms of gout. These crystals are deposited in joints (recall that the classic symptom of gout is an inflamed toe).
Adenosine is degraded in a two‐step reaction. First, the enzyme adenosine deaminase acts on AMP or adenosine nucleoside to yield IMP or inosine.
IMP is cleaved by phosphorolysis of the nucleoside to yield hypoxanthine and ribose‐1‐phosphate. (This reaction is similar to the phosphorolysis of glycogen by glycogen phosphorylase.)
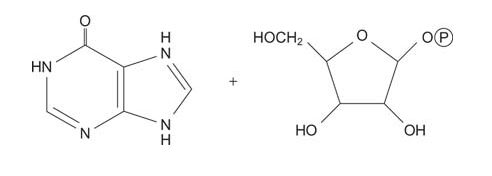
![]()
Guanosine is degraded in a two‐step reaction sequence. First, guanosine phosphorylase phosphorolyses the nucleoside to free guanine and ribose‐1‐phosphate.
![]()
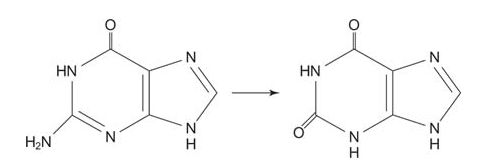
The next reaction is the deamination of guanosine to xanthine. Xanthine needs only one more oxygen to form uric acid.
Xanthine oxidase oxidizes hypoxanthine and xanthine to uric acid, using molecular oxygen, O 2.

![]()
As mentioned earlier, uric acid is only slightly soluble and individuals with impaired secretion or excess production of uric acid are subject to the pain of gout as uric acid precipitates in the joints. Most cases of gout are probably due to impaired excretion of uric acid because of poor kidney function. Because the concentration of uric acid in the blood is near the solubility limit, only a slight impairment of elimination can push the concentration high enough to precipitate uric acid. More frequently nowadays, gout appears in persons whose kidney function is impaired with age, although it is also found in individuals with genetic deficiencies in the level of hypoxanthine‐guanine phosphoribosyl transferase. In the latter case, the salvage pathway does not function well, and more purines must be eliminated through their conversion to uric acid.
The drug allopurinol, which is an inhibitor of xanthine oxidase, effectively treats gout. Allopurinol is structurally similar to hypoxanthine, except that the 5‐membered ring has the positions of the carbon and nitrogens reversed.
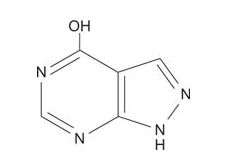
![]()
Xanthine oxidase is able to bind allopurinol and catalyze one oxidation, converting it to a compound that is similar to xanthine. However, after that conversion, the enzyme is trapped in an inactive oxidation state and can't carry out its normal function of forming uric acid. Additionally, allopurinol inhibits the de novo (new, from other compounds; not recycled) synthesis of purines, further decreasing the amount of uric acid formed in the blood.